top of page
SINGLE PARTICLE MICROSCOPY
Understanding the atomic structures of colloidal inorganic nanoparticles and their changes over time is of great importance, as it allows us to predict the physical and chemical properties of the nanoparticle. However, owing to the intrinsic heterogeneity among nanoparticles and the practical difficulty of analyzing nanoparticles in their native liquid phase, the static and dynamic structures of colloidal nanoparticles are hardly accessed. To tackle these obstacles, we are developing experimental and computational methods to study the full 3D structures of single nanoparticles in liquid and their transformations over time.
Based on our original method, named 3D SINGLE (3D Structure Identification of Nanoparticles by GLC EM) or Brownian one-particle reconstruction, we study the 3D structures and dynamics of various metal and semiconductor nanoparticles. We also develop computational methods such as neural-network-based image data processing, and computational chemistry for nanoparticles in realistic environment, as complementary methods to overcome finite spatial and temporal resolutions in 3D SINGLE and to understand nanoparticle dynamics in detail.
Learn more
Multiscale 3D Structure Analysis for Single Nanoparticles
The 3D SINGLE method is a 3D structure determination method based on the free rotation of nanoparticles by Brownian motion in a graphene liquid cell (GLC). Snapshots of a freely-rotating nanoparticle are acquired as a series of 2D images from various projection directions, and 3D structures of the nanoparticle are reconstructed from the projections. For example, by using the 3D SINGLE method, we have obtained 3D atomic structures of Pt nanoparticles with high precision (~19 pm). The 3D SINGLE method allows us to observe unique structural properties of nanoparticles in solution phase. We also develop this method such that it can be used to determine 3D structures of complex structured nanoparticles such as ones with multi-element composition including alloy catalyst nanoparticles and quantum dots.
The core idea of the 3D SINGLE method also works for rotating nanoparticles in various liquid cells, not just for the GLC, imaged in various magnifications. 3D morphologies of nanoparticles can be efficiently characterized by 3D SINGLE using low magnification images, enabling direct observation of size, shape, core-shell interface, and dislocations of individual nanoparticles in 3D. We aim to implement this algorithm into our pipeline for multi-scale 3D structure TEM analysis, in order to resolve 3D structural information of nanoparticles in a high-throughput manner.

Surface Atomic Chemistry of Single Nanoparticles
Since atomic arrangement of a nanoparticle determines its physicochemical characteristics, extensive structural analysis is crucial to systematically understand and exploit its unique properties. Our research focuses on elucidating the structure-property relationships of various nanoparticles through the development of new analytical methods utilizing experimentally resolved 3D atomic structures. Specifically, we construct 3D maps of coordination number (CN) and generalized CN (GCN), which enable us to quantify the surface structures and correlate them to the thermodynamics and catalytic activity of the nanoparticle. We have revealed that sub-3-nm synthesized nanoparticles are enclosed by islands with nonuniform shapes, which leads to complex surface structures. In addition, we implement machine-learning-based DFT calculations based on realistic 3D atomic structures to reveal ligand-surface adsorption chemistry and catalytic activity of nanoparticles in a level considering entire constituent surface atoms. With the combination of those methods, we ultimately aim to resolve the atomic structures of active sites, their distribution on the nanoparticles and their physicochemical properties under catalytic reaction conditions.

Time-resolved 3D reconstruction
We are developing an experimental approach to reconstruct time-resolved 3D atomic structures of a single colloidal nanoparticle in solution and applying it to investigate the nucleation, growth, etching, and phase transitions of individual nanoparticles in 3D atomic resolution. This is done by expanding the 3D SINGLE method to reconstruct time-resolved 3D structures of a nanoparticle undergoing changes in solution. To achieve this goal, we develop the advanced 3D reconstruction algorithm that can produce continuous 3D density maps of the changing single nanoparticle in a time resolution desired for capturing critical events underlying in a phase transition of interest. We also develop a new imaging method where in situ TEM movies of nanoparticles undergoing structural transitions in a graphene liquid cell are captured over tens of seconds with high spatial and temporal resolution.
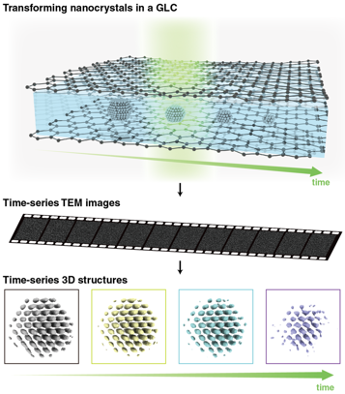
Self-Supervised Image Processing for Advanced In Situ TEM
TEM image series obtained from advanced electron microscopy suffer from different types of noise that prohibits successful information extraction. To solve this problem, we develop a machine-learning based denoising algorithm which can be widely applicable in TEM imaging including, but not limited to liquid cell TEM, time-series STEM, and cryo-TEM. The machine learning-based denoising procedure we are developing aims for not only improving the visual quality of in situ TEM images but also enabling efficient mining of unbiased information without compromising spatial and temporal resolution, leading to the successful down-stream image data process for 3D reconstruction, tomography, and atom-tracking. The method works effectively for a wide range of fields dealing with visual data beyond electron microscopy.

Advanced Graphene Liquid Cell TEM with Reaction Controls
Graphene as windows for liquid cell TEM is advantageous due to its electron transparency and high conductivity, which enables high-resolution imaging. However, controlling liquid geometry or applying external stimulus to the GLC environment is challenging, limiting its applications. To overcome these obstacles, we are developing new types of graphene liquid cells using fabrication techniques such as MEMS to control liquid geometry (location and thickness) and to integrate methods that induce reaction controls (ohm heaters and electrodes). We have developed silicon-based microwell-type chips and nanoarray-type anodic aluminum oxide-based GLCs which enable quantitative analysis of reactions occurring within liquid pockets of controlled geometries. Furthermore, we are developing silicon-based chips with windows made of graphene capable of operando microscopy of single nanoparticles in liquid.


Molecular Dynamics for Nanoparticle Atomic Structures
We are working on molecular dynamics (MD) simulation to observe reactions of nanoparticles in realistic condition. It will predict various pathways governing diverse phase transitions of nanoparticles. In addition, it will also complement our liquid phase TEM observation of nanoparticle phase transitions by improving temporal resolution to overcome experimental limit. We implement new sampling methods to enhance calculation performance, allowing us to observe reactions at longer time scales compared to conventional methods. Specifically, we make use of the weighted ensemble (WE) method, an unbiased sampling strategy, that keeps track of important kinetic information. With this method, we investigate the nucleation, growth, and etching processes of nanoparticles at atomic scale.
bottom of page